Back from BlueHats, satellite to OSX 2022
GNU Guix, toward the computational reproducibility?
Note: BlueHats is an initiative to gather people – from staff employed by French state to volunteer citizens – who are contributing to free software in use by French public administration. All opinions are mine.
The BlueHats event is satellite to Open Source Experience (OSX22, ex-Paris Open Source Summit). The day was organized by the team of Bastien Guerry working about commons. I presented Guix and how it helps in « Open Science ». Back in 2019, Guix had already been presented in a previous edition.
The talk was in French with a slot of 15-20 minutes, questions included. The slot was very short considering the heterogeneity of the audience; far from day-to-day scientific production. Similarly to the previous edition, I have tried to explain what is the « Reproducible Research » challenge in the modern age of data and computing. The slides (in French) are available here (PDF) (source). Let recap for non-French speaker!
The context is the production of a scientific result depending on some computations. The main question is summarized by: why does it work for Alice and not for Carole? and vice-versa. How Alice and Carole can they observe the same output from the same input?
Note: Using another frame but expanded, auto-promotion with the article Toward practical transparent verifiable and long-term reproducible research using Guix published in Nature Scientfic Data (2022).
the message you should get back to home (I hope)
Somehow, the world is becoming open, isn’t it? Now, we have open journal, open data, open source, open science, well open something. People are working hard to make this « open world » a reality and, despite this « openness », the replication crisis is still there. Speaking about a computation, what is the problem about reproducibility in a scientific context? (Even if all is becoming open.)
What is a « computer »? It is an instrument that allows to automatically deal with data. Assuming the analogy that a computation is similar to a measurement, then the experimental conditions / setup of this measurement by that instrument is similarly captured by the computational environment. How do we control this computational environment?
Consider this simple example: the computation of some Bessel function in the
language C.
#include <stdio.h> #include <math.h> int main(){ printf("%E\n", j0f(0x1.33d152p+1f)); }
Nothing fancy! However, Alice and Carole do not see the same result:
5.643440E-08 vs 5.963430E-08. Why? The code is open and available though.
If the difference is significant or not is let to the scientific valuation of
the field – depending on the context where this computation would be
involved.
Random questions around this computation:
- what is the
Ccompiler? - which libraries (
stdio.handmath.h)? - which versions?
- which compilation’s option?
- etc.
For instance, Alice and Carole are running the same C compiler: « GCC at the
version 11.2.0». Why is different so? Answer: Because they do not use the
same compilation option; Alice is running the default when Carole is running
-lm -fno-builtin. Damned constant folding compiler optimization. :-)
The language (here C) does not matter and similar examples about Python, R,
Ruby, Perl, etc. are easy to spot. Do not miss the core point here: Alice and
Carole are running their computation in two different computational
environments. Despite the source code is available and they agree on version
labels, they do not get the same result. The bold conclusion is: open source
and version label is already good but far to be enough to be able to
reproduce a computation! In short, version labels capture only a tiny part of
the computational environment.
Now we have identified that the computational environment is important, the questions around are:
- what are the source for each software?
- which software is required for building?
- which software is required for running?
- how each software is it produced itself? And recursively.
A precise answer to these questions means a fine control of the variability
of the computational environment. Therefore, how do we capture these
information? Well, the standard answers range from package manager as apt,
Brew, Conda, etc. to containers as Docker, Singularity, etc. or even module
files. From my point of view, these answers are individually not enough.
Let reword the situation about the reproducibility in scientific context:
- From the « scientific method » point of view: all the issue is about controlling what could be variable from one computational environment to another.
- From the « scientific knowledge » point of view: all the issue is to
collect universal – as much as possible – knowledge, which means:
- two independent observers must be able to get the same result,
- the observation must be sustainable – as much as possible.
Therefore, in a world where (almost) all is numerical data, the core question about reproducibility in scientific context is: how to redo later and overthere what had been done today and here? (implicitly using a « computer »). “Later” and “overthere“ mean running the computation from today on that machine using another machine in the future (from 6 months to 5 years later, say!).
where Guix is helping (I estimate)
As underlined above, the standard solutions are:
- package manager:
apt(Debian/Ubuntu),yum(Redhat), etc. - environment manager: Conda,
modulefiles, etc. - container: Docker, Singularity, etc.
Guix is an elegant solution that provides all these 3 solutions in one and tackles all the individual annoyances of each of the others. Guix is computational environment manager on steroids:
- package manager (as
apt,yum, etc.) - transactional and declarative (rollback, concurrent versions, etc.)
- producing redistributable pack (as Docker or Singularity container)
- also producing isolated virtual machines (as Ansible or Packer)
- …one yet another GNU/Linux distribution…
- last, a Scheme library (allowing powerful extensions)
Guix runs on the top any Linux distribution without interfering with this host Linux distribution. It is easy and painless to try… :-) See the Bash shell script and the instructions, especially do not miss the note.
I will not present here all the nice features provided by Guix. Let focus only on one key design explaining why I think Guix is a game changer for computing reproducibility in scientific context.
Above, we were considering the situation where Alice says: « GCC at the
version 11.2.0 ». What does it mean exactly? GCC is a C compiler and it
requires other software to be built and run. In a nutshell, the graph of
dependencies of « GCC at the version 11.2.0 » looks like,
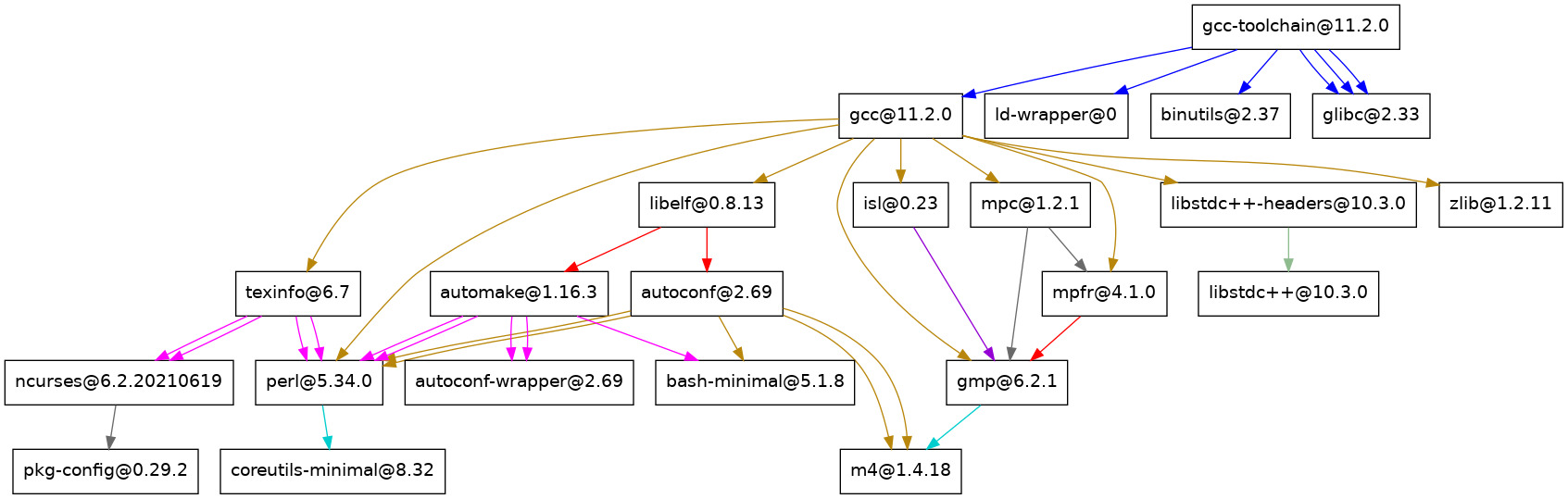
where it means that each square box (node of the graph) is a dependency in
order to get « GCC at the version 11.2.0 ». As you can see, one dependency is
the software mpfr at the version 4.1.0. Now, if this node mpfr@4.1.0 is
instead replaced by mpfr at the version 4.0, do we get the same binary version
of GCC? If Alice and Carole do not see the same result for one computation,
is it because of a flaw in their scientific study or is it a variation with
the computational environment? If they cannot deeply scrutinize all the
computational environment, from the source code to the compilation option of
each node, how can they verify where the difference comes from?
For more details, give a look at this other post considering the situation about the R programming language. All apply equally.
When Alice says « GCC at the version 11.2.0 », behind this benign claim is hidden a graph. And that graph of software dependencies can be large: for instance, 915 dependencies (nodes) for building and running the plotting Python library Matplotlib, and 1439 dependencies for the scientific Python library SciPy. It is not affordable to manually list all the dependencies; aside that the various options for compiling and configuring the dependencies are also required.
Guix is able to capture the graph using the command guix describe. This
command returns the exact state of the graph of dependencies. This state is
summarized by a file containing a list of repositories and their exact state
(URL location, branch name, commit hash). And these repositories contain
package definitions. Therefore, that file captures all the graph, and knowing
it, Guix is able to instantiate the exact same state, whatever where and
whenever.
Let fix the idea with an example. For instance consider,
$ guix describe
Generation 76 Apr 25 2022 12:44:37 (current)
guix eb34ff1
repository URL: https://git.savannah.gnu.org/git/guix.git
branch: master
commit: eb34ff16cc9038880e87e1a58a93331fca37ad92
It means that the current revision (state) is using only one repository
(channel) which is the default one. Therefore, in this case, the hash eb34ff1
captures all the graph of dependencies. Other said, when Alice says « Guix at
the revision eb34ff1 », then Carol knows all the information to rebuild the
exact same computational environment.
In a few words, the workflow for a collaboration is:
Alice describes her computational environment, by:
a) listing all the software she needs, using the file
list-tools.scmand she generates her computational environment, running, e.g.,$ guix shell -m lists-tools.scm
b) but that is not enough. She also needs to provide the revision (state), of Guix itself and also potentially some other channels. She runs,
$ guix describe -f channels > state-alice.scm
Last, Alice shares her computational environment by sharing the two files:
list-tools.scmandstate-alice.scm.Carole builds the exact same environment as Alice using the two Alice’s files,
guix time-machine -C state-alice.scm -- shell -m list-tools.scm
It is where Guix shines! It is really straightforward to share computational environment by only sharing two files. Obviously, these files can be tracked with the project itself with any version control system as Git.
At this point, it addresses the part « run the same thing elsewhere » but what about « run later ». Internet (extrinsic URL identifier) is ephemeral and the link-rot is one strong concern for reproducibility in scientific context. What are the guarantees that the source code hosted on popular forges (say as GitHub or Gitlab or else) will be still there some time later? Example of past popular forges: Google Code (closed in 2016), Alioth (from Debian replaced by Salsa in 2018), Gna! (closed in 2017 after 13 years of intensive activities), Gitorious (closed in 2015 being at some point the second most popular hosting Git service), etc.
Please Software Heritage, the floor is yours! Software Heritage collects, preserves and shared software in source code form, because, quoting their website, software embodies our technical and scientific knowledge and humanity cannot afford the risk of losing it. Software Heritage try hard to guarantee the preservation of source code in the very long term.
Again, it is where Guix shines! Unique to my knowledge. Whatever the status of the original location of the source code, if it had been archived in Software Heritage, then Guix is able to fallback – from channels (package definition) to software source code. For this fallback in action, give a look at this example.
closing words (I guess)
Reproducibility in scientific context is about how to redo later and overthere what had been done today and here. It implies track and transparency. Transparent tracking allows to study bug-to-bug, i.e., to deeply understand the relationship between inputs and outputs, between cause and effect – the pillar of scientific knowledge.
Guix should manage everything when using,
guix time-machine -C state.scm -- <cmd> -m tools.scm
if it is specified:
- how to build (
state.scm), - what to build (
tools.scm).
Convinced by Guix, right? Join the fun, join the initiative GuixHPC1!
Footnotes:
HPC meaning all scientific computing, not only run on BIG cluster. :-)
